A High-Throughput Cell Analysis System to Determine Chromosome Territory Boundaries and Spatial Organization
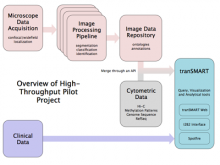
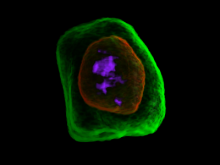
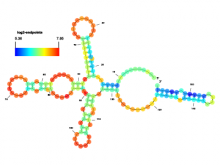
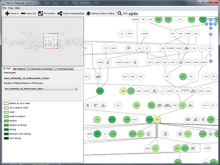
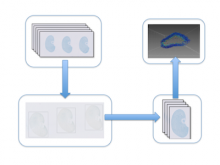
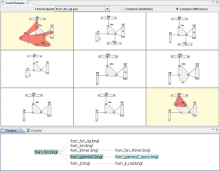
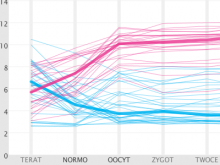
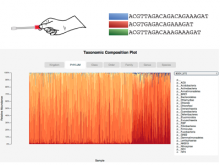
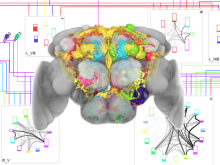
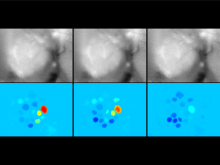
Researchers at the University of Michigan, in collaboration with the Brady Institute at Johns Hopkins University, have begun development of a high-throughput, semi-automated cell analysis system to characterize a number of cellular features including chromosome territory boundaries and their three-dimensional arrangement within the cell nucleus and to correlate these features to prostate disease. Image data will be acquired using a combination of standard confocal and widefield microscopy along with super-resolution imaging techniques to refine chromosome territories and to identify sub-territories. Using the Taverna framework, an image processing pipeline is being developed to facilitate tasks such as channel separation, interest operations and classification, spectral unmixing, clustering, data fusion, feature extraction, and visualization across large populations of cells. The pipeline framework will record provenance for all of the processing modules. Additionally, microscope vendor metadata will be saved along with user annotations that describe data files and the relationships between those files. For example, user supplied metadata may include experimental conditions, fluorophores, etc. We currently support user annotations from the Cell Ontology and the Biological Imaging Methods Ontology, and are actively investigating other ontologies in the NCBO BioPortal. Resulting microscopy data will be stored in an Omero image repository and will be merged with cytometric data through an API to tranSMART, a data sharing and analytics platform for translational biomedical research. The key visualization challenge for this project is the data fusion between multi-resolution microscopy modalities along with cytometric data such as Hi-C and methylation data.
BioVis 2013 Information