HitWalker2: An interactive and queryable web-based framework for variant prioritization in precision medicine
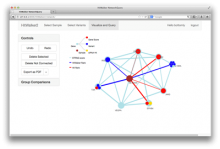
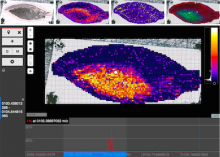
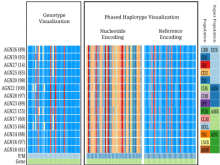
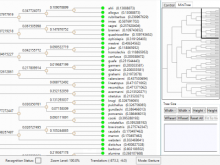
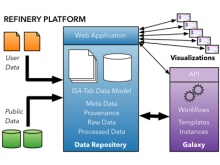
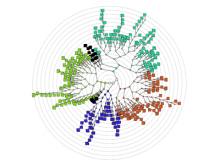
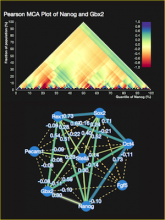
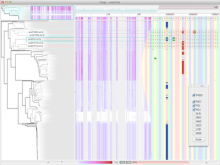
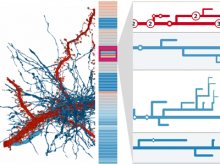
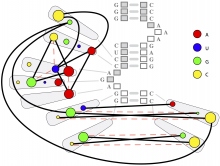
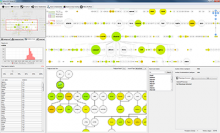
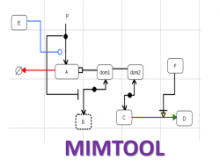
One approach to determining the causal/driver variants for individual cancer patients is the prioritization of DNA variants derived from whole exome or genome sequencing relative to the functional results of other lab based assays such as drug sensitivity or siRNA screens. The network-based prioritization and visualization methods implemented in the HitWalker R package was developed to meet this need (Bottomly et al 2013). However, the original HitWalker framework required R proficiency for customization and produced static images and tables. HitWalker2 was developed to provide a more dynamic and user-friendly experience to allow utilization by diverse groups of users. A web-based framework extends the functionality of HitWalker providing the ability to interact with the graphical network output and ask intuitive questions of the genes and samples contained within the database. The user can specify groups of genes or samples to retrieve basic summary statistics for the groups as well as look for common features (e.g. samples with specific disease types or genes with specific siRNA hits) within or between groups. Additional visualizations are being implemented such as those displaying expression outliers and aberrant expression from RNASeq and the lab assay data. Our current repository contains whole exome variant and functional assay data from a moderate sized cohort of 500 samples. HitWalker2 will be made publically available to academic users. As the framework is accessible to both clinicians and researchers facilitating robust computational prioritizations and querying capability, HitWalker2 will provide an effective means to facilitate precision medicine research.
BioVis 2014 Information