An interactive visualisation tool for the hierarchical clustering of large data sets
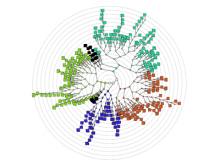
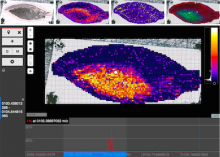
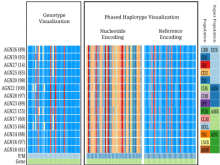
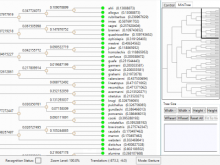
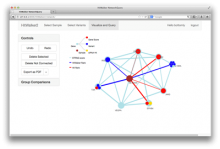
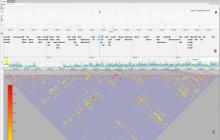
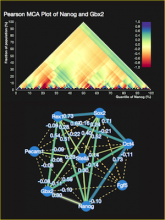
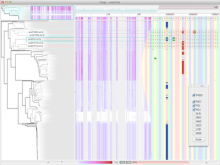
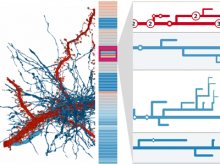
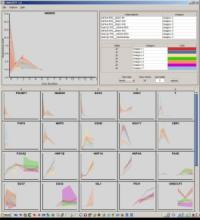
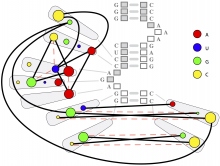
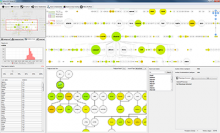
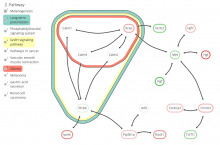
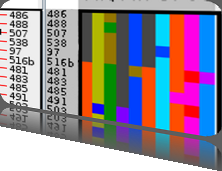
Rigid adherence to pre-specified thresholds and static graphical presentations can lead to incorrect decisions on merging of clusters. We have developed an interactive approach to visualize progress of the average linkage clustering algorithm. The user can see similarities within the clusters and edit them if appropriate. An interactive dendrogram is constructed, which can be displayed in different layout configurations and scales. Results can be viewed in real time, when the user modifies parameters such as the similarity threshold at which clusters are merged and the level of tolerance to intra-cluster distinctiveness, for which we use the term 'heterogeneity'.
BioVis 2014 Information