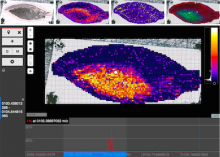
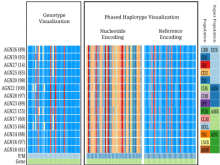
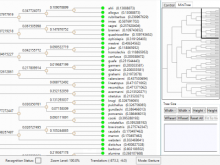
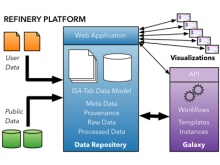
Visualization of gene expression information within the context of the mouse anatomy
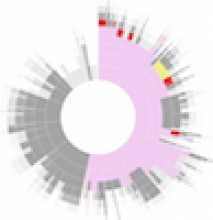
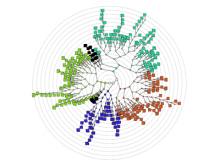
Background: The eMouse Atlas of Gene Expression (EMAGE) is an online resource that publishes the results of in situ hybridisation gene expression experiments on the developmental mouse. The resource provides comprehensive search facilities, but few analytical tools or visual mechanisms for navigating the data set. To deal with the missing visual navigation, this paper explores the application of sunburst and icicle visualizations within EMAGE.
Results: A prototype solution delivered a simple sources. It is increasingly clear that users require assistance when navigating, searching and understanding these information silos.
Conclusions: The sunburst and icicle visualizations have been shown to be effective tools for summarising gene expression data. The sunburst with its space saving radial layout was found especially useful for providing an overview of gene families or pathways. Work is ongoing to integrate these visualizations into EMAGE.
BioVis 2014 Information