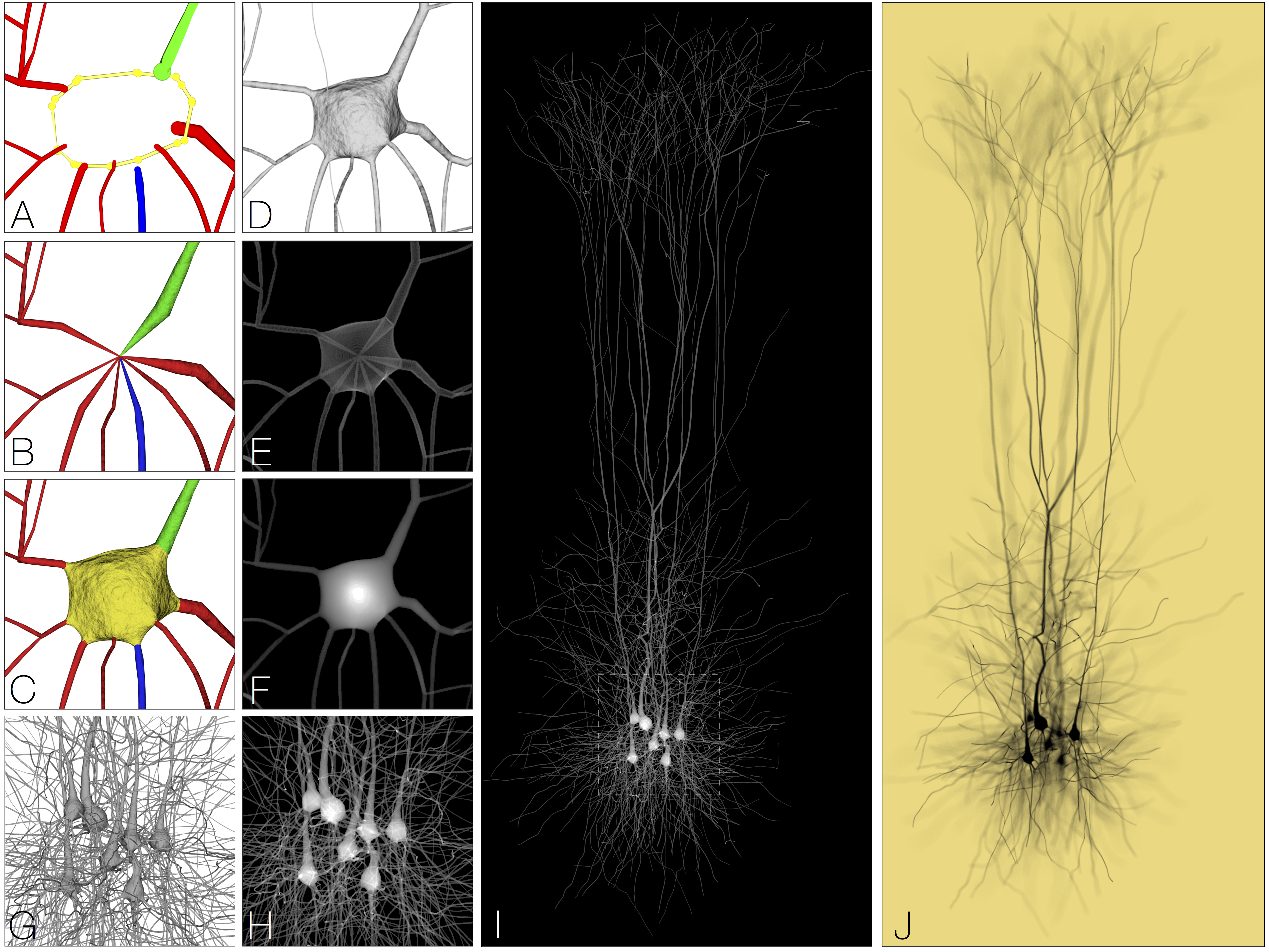
Accepted Papers for BioVis@ISMB 2017
BioVis@ISMB 2017 has now concluded
Check back next year for more opportunities to participate in BioVis@ISMB. We also have a workshop being held in conjunction with IEEE VIS, BioVis@VIS, that will take place in October 2017.
All accepted papers are accessible via BMC Bioinformatics
|
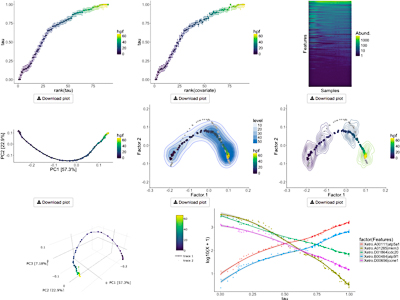
Bayesian Unidimensional Scaling for visualizing uncertainty in high dimensional datasets with latent ordering of observations
Lan Huong Nguyen* and Susan Holmes Presentation Info: "Machine Learning and Medicine" session between 16:30 to 16:50 Background: Detecting patterns in high-dimensional multivariate datasets is non-trivial. Clustering and dimensionality reduction techniques often help in discerning inherent structures. In biological datasets such as microbial community composition or gene expression data, observations can be generated from a continuous process, often unknown. Estimating data points' `natural ordering' and their corresponding uncertainties can help researchers draw insights about the mechanisms involved. Results: We introduce a Bayesian Unidimensional Scaling (BUDS) technique which extracts dominant sources of variation in high dimensional datasets and produces their visual data summaries, facilitating the exploration of a hidden continuum. The method maps multivariate data points to latent one-dimensional coordinates along their underlying trajectory, and provides estimated uncertainty bounds. By statistically modeling dissimilarities and applying a DiSTATIS method to their posterior samples, we are able to incorporate visualizations of uncertainties in the estimated data trajectory across different regions using confidence contours for individual data points. We also illustrate the estimated overall data density across different areas by including density clouds. One-dimensional coordinates recovered by BUDS help researchers discover sample attributes or covariates that are factors driving the main variability in a dataset. We demonstrated usefulness and accuracy of BUDS on a set of published microbiome 16S and single cell RNA-seq data. Conclusions: Our method effectively recovers and visualizes natural orderings present in datasets. Automatic visualization tools for data exploration and analysis are available at: https://github.com/nlhuong/visTrajectory |