|
Paper: Interactive Exploration of Ligand Transportation through Protein Tunnels
Katarina Furmanova*, Miroslava Jaresova, Jan Byska, Adam Jurcik, Julius Parulek, Helwig Hauser, Barbora Kozlikova
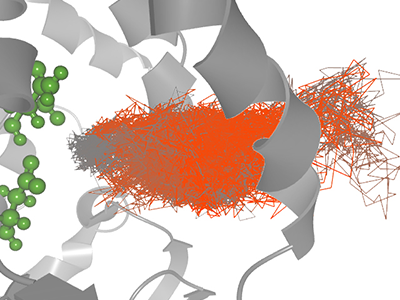
Background: Protein structures and their interaction with ligands have been in the focus of biochemistry and structural biology research for decades. The transportation of ligand into the protein active site is often complex process, driven by geometric and physico-chemical properties, which renders the ligand path full of jitter and impasses. This prevents understanding of the ligand transportation and reasoning behind its behavior along the path. Results: To address the needs of the domain experts we design an explorative visualization solution based on a multi-scale simplification model. It helps to navigate the user to the most interesting parts of the ligand trajectory by exploring different attributes of the ligand and its movement, such as its distance to the active site, changes of amino acids lining the ligand, or ligand "stuckness". The process is supported by three linked views -- 3D representation of the simplified trajectory, scatterplot matrix, and bar charts with line representation of ligand-lining amino acids. Conclusions: The usage of our tool is demonstrated on molecular dynamics simulations provided by the domain experts. The tool was tested by the domain experts from protein engineering and the results confirm that it helps to navigate the user to the most interesting parts of the ligand trajectory and to understand the ligand behavior.
|