|
Poster: Co-visualisation of Close Genetic Relatedness of Mycobacterium Tuberculosis Isolates with Complex Meta-Data
Trien V. Do, Oriol Mazariegos Canellas, Derrick Crook, Tim Peto, David Wyllie
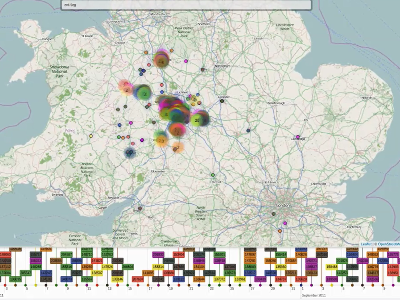
Routine DNA sequencing of all Mycobacterium tuberculosis samples isolated in England is projected to be operational within the next 12 months. We have recently created a web application which allows identification of closely genetically related samples and storage of meta-data about the individuals from whom they were obtained. This capability is of interest since breaking chains of transmission depends on their rapid identification and pairs of individuals with highly related samples are more likely to have transmitted to each other. However, prioritising interventions requires expert review of the genetic relationships, intervals between isolation, location, and individual risk factors, inter alia. This paper presents a visualisation approach to address this issue. A user (e.g. a public health doctor, or TB control nurse) logs into the web application then can select sets of individuals of interest, either by date, name, specimen number, or genetic relatedness to all cases. In 'set of samples' mode, the application allows residential addresses, times of isolation, and genetic relationships to be co-visualised for a sample set in a map view and a timeline. In 'single case exploration' mode, links between a single case and others can be displayed, with metadata displayed in tooltip like format (e.g. name, age, address, history, resistance, contact number). The user can 'Explore from here', upon which the current sample will become a new target sample. Thus, iterative exploration of transmission networks is possible. This application is being co-developed and evaluated between the University of Oxford and Public Health England TB control professionals.
|