Glom: Image Segmentation for Glomerular Counting
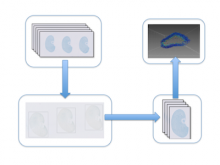
Precisely counting the number of glomeruli is a critical step to understand kidney disease development patterns. High throughput microscopy imaging systems can produce kidney histological section images automatically. However, to apply image registration and 3D cell counting algorithms on image stacks, multiple sections on one or multiple slides need to be cropped out and saved as one image stack. \ \ To assist quantifying glomeruli phenotypes, we have developed an automated system to add bounding rectangles to outline the histological sections on each slide and convert those sections into one image stack. Our approach applies a discrete Laplace filter to highlight cellular texture, followed by a Gaussian blur filter to remove possible system artifacts. Finally, Li�۪s Minimum Cross Entropy threshold algorithm is used to add bounding rectangles to highlight related histological sections. \ \ Artifacts from system errors can lead to false negative or false positive bounding rectangles, which need a significant amount of downstream work to correct. To reduce error rates, we have also developed an interactive visualization interface that allows users to quickly examine the processed images to make necessary adjustments to the bounding rectangles. Users can edit, add and remove the bounding rectangles before the images are cropped for downstream processing. \ \ With both automated cell histological section detection and a visualization tool for bounding box adjustment, we can quickly and accurately quantify the glomeruli from an experiment. Our approach allows more efficient and accurate image recognition of biologically related objects, which is an indispensible step in a developmental biology study. \