A Journaling System for Rule-Based Biochemical Models
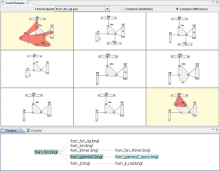
Rule-based modeling allows for the efficient construction and simulation of models representing biochemical interactions within cells. As systems biologists iteratively develop these models, tracking alterations of the model becomes difficult. Additionally, as parent models are adapted for particular purposes, it is easy to lose track of model versions and the assumptions made at various branch points. A system to interactively browse the structure of a family of models, as well as identify any parent or child of each model, would greatly facilitate model development and analysis. This poster highlights the development of such a journaling system. A small multiples view displays the contact map rendering, which displays molecular components and interactions, of each model in the development family. A layout stabilization mechanism allows for each model to be snapped into a common layout for better visual comparison. Models are sorted by similarity to the most complete model. Users can interactively compare the similarities and differences between pairs of models. A history tree view shows the development of the model family with respect to time. Additionally, a list of the model simulations, as well as the model structure at each simulation, is linked to each node in the tree. This journaling system is built on top of the BioNetGen language and simulator. Informal feedback from biology collaborators has shown great interest in this approach. Some of the suggested potential applications range from visually identifying locations for combining models to identifying a core structure within a family of models.