Novel approach to modelling protein-protein interactions in biological space
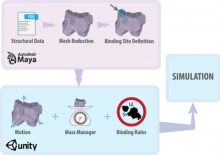
The main aim of this work was to develop an accurate and detailed method to simulate protein-protein interactions and the formation of large complexes, and use it to study the properties of Post Synaptic Density (PSD) formation in a dendritic spine. The methodology is based on simulating accurate protein geometries diffusing in space and interacting only via their binding sites. Coarse grained representation of the protein surfaces were produced starting from their PDB file structure, using Autodesk Maya. Subsequently, Unity, a game engine was used to simulate diffusion and reaction of the proteins models in the simulation environment. Diffusion in 2/3D was implemented as mass driven, with explicit calculations of translation and rotation, and the 2D diffusion was implemented to be independent of the shape of the container, its concave nature, and its tessellation. The protein binding strategy was implemented based on collision theory. The method was fully validated for the 2D and 3D diffusion and for complex formation, and applied to the study of the Post-Synaptic Density (a proteinaceous organelle present on the membrane of excitatory spines) assembly formation.





