Active Data Canvas: linking data to knowledge
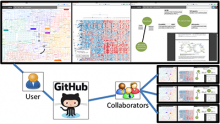
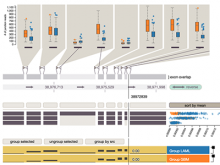
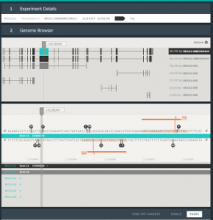
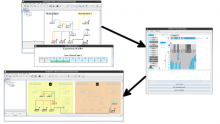
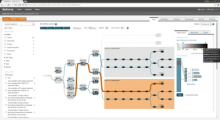
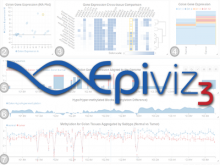
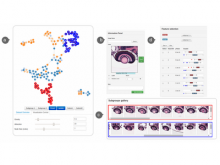
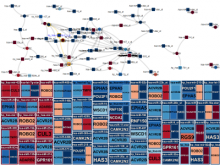
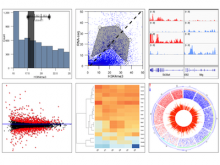
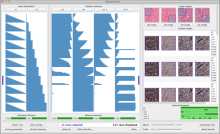
Intuitive data visualization is critical for biomedical data analysis; especially essential is the ability for non-computational scientists to productively browse the data and apply their domain knowledge. We present Active Data Canvas, a web-based visual analytic tool suite that allows dynamic data interaction in familiar and intuitive contexts, like pathways and heatmaps. Users can track emerging hypotheses by pinning data to the Canvas, a Pinterest style board for aggregating and assimilating thoughts. As data are pinned, the Canvas proactively searches structured and unstructured knowledge bases (e.g. KEGG, Pubmed, Wikipedia etc.) to fetch relevant information. In this manner it becomes a productive digital assistant, lessening the burden on the researcher and freeing time for contemplative analysis. The Active Data Canvas promotes collaboration by using GitHub as the back-end data store. Via the ambitious task of versioning data, analyses and emergent hypotheses, users who collaborate on a project can share discoveries in real time. We used the Active Data Canvas to analyze ovarian cancer data for 174 tumors. After quality control and initial analysis, the R object was directly imported along with the clinical metadata. New hypotheses and conclusions were discovered via the interaction with the Active Data Canvas that had not been discovered in the months of analysis via statistical programming scripts.
BioVis 2015 Information