Integrated visual analysis of protein structures, sequences, and feature data
Background: To understand the molecular mechanisms that give rise to a protein's function, biologists often need to (i) find and access all related atomic-resolution 3D structures, and (ii) map sequence-based features (e.g., domains, single-nucleotide polymorphisms, post-translational modifications) onto these structures.
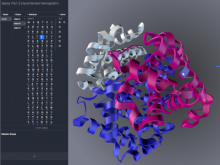
Results: To streamline these processes we recently developed Aquaria, a resource offering unprecedented access to protein structure information based on an all-against-all comparison of SwissProt and PDB sequences. In this work, we provide a requirements analysis for several frequently occuring tasks in molecular biology and describe how design choices in Aquaria meet these requirements. Finally, we show how the interface can be used to explore features of a protein and gain biologically meaningful insights in two case studies conducted by domain experts.
Conclusions: The user interface design of Aquaria enables biologists to gain unprecedented access to molecular structures and simplifies the generation of insight. The tasks involved in mapping sequence features onto structures can be conducted easier and faster using Aquaria.
BioVis 2015 Information