A new hierarchical 3D random walk visualization for whole chromosome sequences
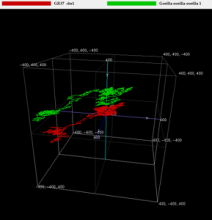
Graphical techniques are a very powerful tool for the visualization and analysis of long DNA sequences by providing useful insights into local and global characteristics and the occurrences, variations and repetition of the genomic sequences. Within these methods, the random walk plot is a basic technique to visualize a huge genome on 2D or 3D geometric space. One problem in these geometric random plotting is the loss of genetic information due to the complementary nucleotide base pairs. In this work we propose a new 3D random walk plotting method to suppressing complementary base pair cancelation effect by allowing z-axis relaxation. Since the final “geometric genomes” constructed on 3D space is of similar complexity as fractal objects, it is necessary to simplify them into a manageable data structure. So we give one simplification algorithm for the geometrically visualized long sequences with alpha-hull polygon modeling. So we can compare the geometric similarity of these simplified polygons which are constructed from more than 100 mega base DNA sequences, which enables us to recognize the partial matching and the global similarity more intuitively without using any computation-intensive sequence alignment. Our experiment showed that our method is quite fast and effective in comparing whole chromosomes of more than 10 mammals.
BioVis 2015 Information