Pan-Tetris: an interactive visualisation for Pan-genomes

Background: Large-scale genome projects have paved the way to microbial pan-genome analyses. Pan-genomes describe the union of all genes shared by all members of the species or taxon under investigation. They offer a framework to assess the genomic diversity of a given collection of individual genomes and moreover they help to consolidate gene predictions and annotations. The computation of pan-genomes is often a challenge, and many techniques that use a global alignment-independent approach run the risk of not separating paralogs from orthologs. Also alignment-based approaches which take the gene neighbourhood into account often need additional manual curation of the results. This is quite time consuming and so far there is no visualisation tool available that offers an interactive GUI for the pan-genome to support curating pan- genomic computations or annotations of orthologous genes.
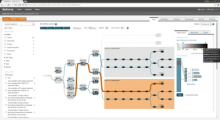
Results: We introduce Pan-Tetris, a Java based interactive software tool that provides a clearly structured and suitable way for the visual inspection of gene occurrences in a pan-genome table. The main features of Pan-Tetris are a standard coordinate based presentation of multiple genomes complemented by easy to use tools compensating for algorithmic weaknesses in the pan-genome generation workflow. We demonstrate an application of Pan-Tetris to the pan-genome of Staphylococcus aureus.
Conclusions: Pan-Tetris is currently the only interactive pan-genome visualisation tool. Pan-Tetris is available from http://bit.ly/1vVxYZT.
BioVis 2015 Information