VisRseq: R-based visual framework for analysis of sequencing data
Background: Several tools have been developed to enable biologists to perform initial browsing and exploration of sequencing data. However the computational tool set for further analyses often requires significant computational expertise to use and many of the biologists with the knowledge needed to interpret these data must rely on programming experts.
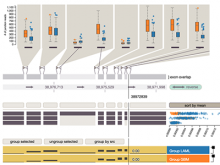
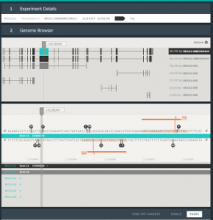
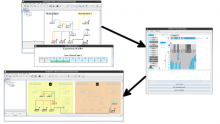
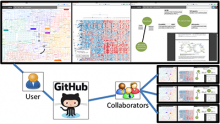
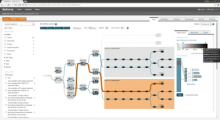
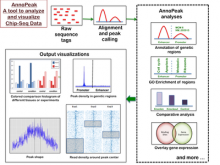
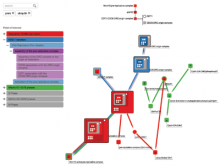
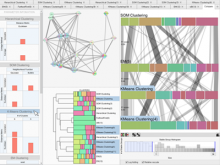
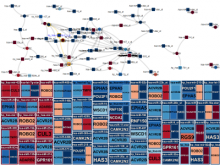
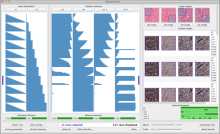
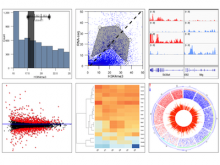
Results: We present VisRseq, a framework for analysis of sequencing datasets that provides a computationally rich and accessible framework for integrative and interactive analyses without requiring programming expertise. We achieve this aim by providing R apps, which offer a semi-auto generated and unified graphical user interface for computational packages in R and repositories such as Bioconductor [1]. To address the interactivity limitation inherent in R libraries, our framework include several native apps that provide exploration and brushing operations as well as an integrated genome browser [2]. The apps can be chained together to create more powerful analysis workflows.
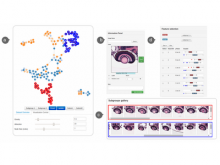
Conclusions: To validate the usability of VisRseq for analysis of sequencing data, we present two case studies performed by our collaborators and report their workflow and insights.
BioVis 2015 Information