BioVis 2011 Abstract
KnomePathways: A Tool to View and Query Gene Interaction Networks in Individual Human Genomes
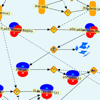
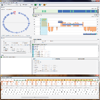
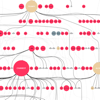
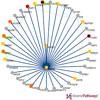
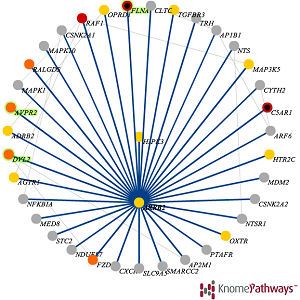
Common diseases and other complex human phenotypes likely trace to interactions between variants in distinct genes. Spotting such interactions is hard, as each person’s genome carries millions of distinctive variants, distributed among thousands of genes. To overcome this challenge, we present KnomePathways, a tool that overlays variants found in individual human genomes onto publicly annotated sets of interacting or co-expressed genes. Using a simple color scheme, rich underlying annotation, and powerful comparative querying, KnomePathways lets users quickly find gene-pairwise interactions that may be functionally disrupted – and, more broadly, networks of genes that are enriched for suspect variants in some genomes (e.g., cases) versus others (e.g., controls).
BioVis 2011 Papers and Abstracts