Bulk Synchronous Visualization
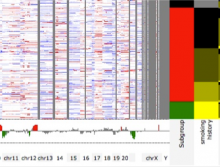
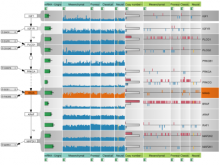
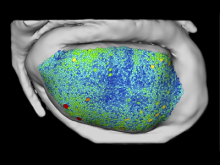
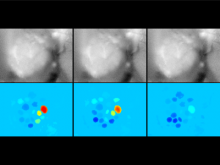
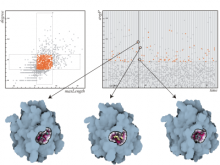
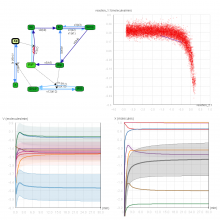
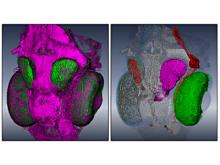
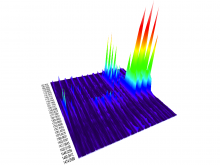
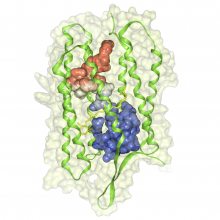
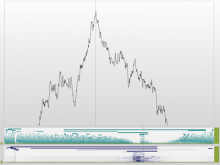
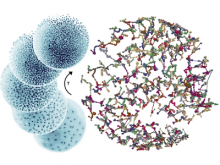
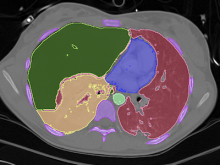
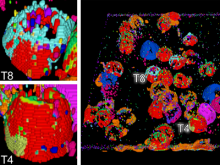
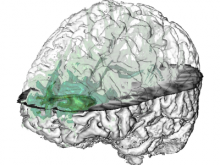
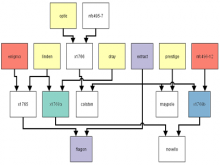
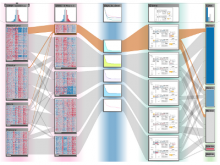
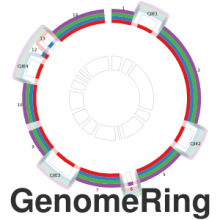
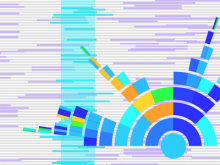
Many biological applications require computationally expensive high resolution visualizations. Current displays have the required resolution, and current computers the required computational resources. However, it is still challenging to write programs that can utilize these resources. We describe the bulk synchronous visualization (BSV) model for interactive parallel visualization. The data to be visualized is split into many parts which are visualized in parallel using hundreds of windows. It makes it easier to write parallel visualizations since there is no multi-threaded programming, and resource management is hidden by the system. The BSV system is work in progress.
BioVis 2012 Information