Can Adjacency Matrices help in the exploration and understanding of Multi-Omics Data?

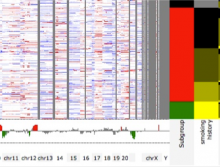
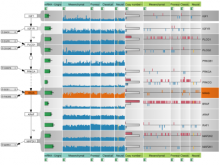
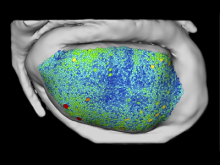
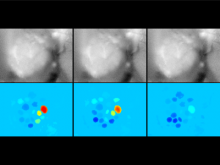
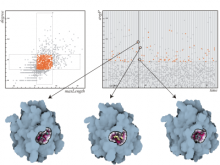
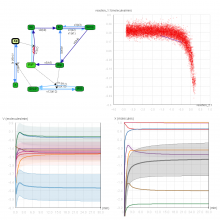
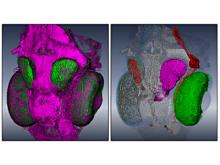
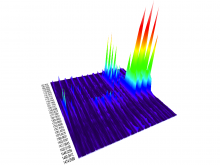
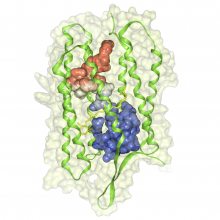
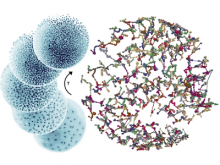
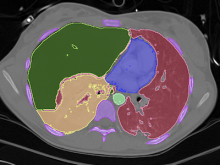
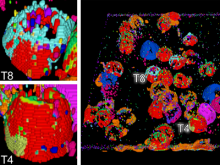
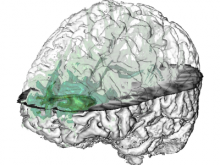
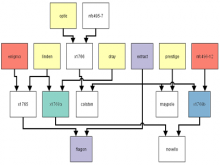
One of the current critical steps of biological science is the hypothesis generation from genome-wide data and the integration of various multi-omics data into a single, coherent view. To facilitate this, we are exploring the possibilities of multiple gene network matrices visualizations and are showing playful ways of interacting with the matrices. The goal is to create an environment, which would enable biologists to obtain insights from large biological multi-dimensional data.
BioVis 2012 Information