GenomeRing: alignment visualization based on SuperGenome coordinates
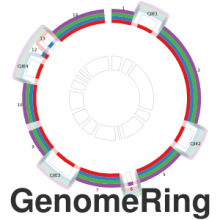
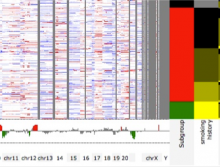
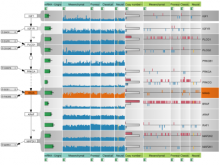
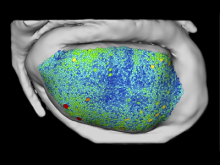
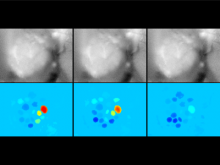
We have developed a visual analytics framework encompassing two complementary approaches to tackle the problem of multiple genome alignment visualization. For a given multiple genome alignment, the SuperGenome algorithm computes a common coordinate system for all genomes in the alignment. With this coordinate system, genome annotations can be placed consistently even in the presence of insertions, deletions, and rearrangements between the genomes. Secondly, based on the SuperGenome coordinate system, GenomeRing visualizes the multiple genome alignment in a circular layout. GenomeRing is a highly interactive visualization tool integrated in our software Mayday.
BioVis 2012 Information