Getting Into Visualization of Large Biological Data Sets
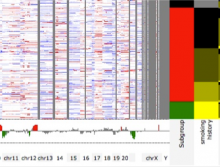
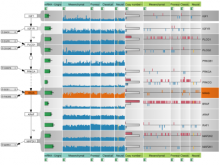
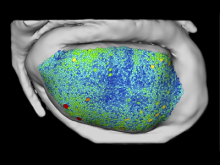
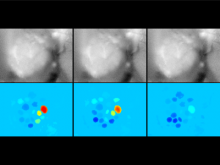
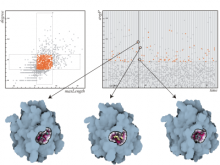
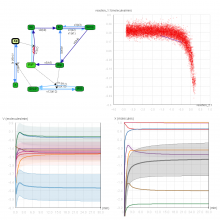
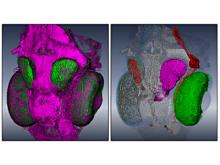
Creating impactful visualizations in biology requires productive dialogue between computer scientists and biologists. The utility and longevity of a visualization method is increased if the system is made independent of specific analysis methods, given flexibility to address a variety of data types and hypotheses and useful for both data exploration and communication. From experience in creating and maintaining Circos, the community standard for visualizing genome comparisons, and Hive Plots, a method for quantitative network visualization, we present strategies to facilitate this multi-disciplinary dialogue and identify critical components of visualizations designed for print.
BioVis 2012 Information