mzRepeat: Visual Analysis of Lipids in Mass Spectrometry
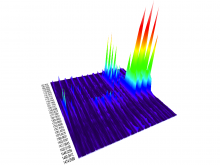
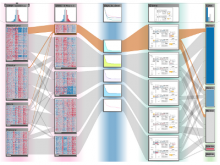
Mass spectrometry is widely used for identifying molecules that have similar structures. A mass spectrum obtain from lipids using MALDI-MS (matrix-assisted laser desorption/ionization-mass spectrometer) was observed to have peaks at equal intervals on the m/z axis. If these repeated peaks can be collectively shown, analysis can be optimized even further. In this study, we propose a visualization system for analyzing repeated observed peaks on the m/z axis. The system, mzRepeat, was developed and evaluated by analyzing butter. A feature of mzRepeat is that ions that differ only in number of repeating structures are shown collectively. \ It is especially effective for analysis of many peaks from the samples that contain several molecular species. \
BioVis 2012 Information