The New UCSC Cancer Genomics Browser
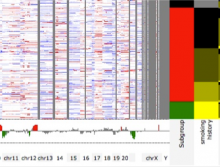
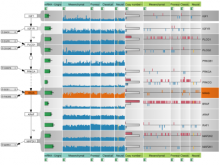
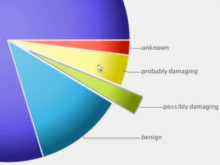
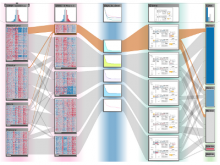
The UCSC Cancer Genomics Browser is a web-based tool to integrate, visualize and analyze cancer genomics and clinical data. Users can order, filter, aggregate, classify, and display data interactively based on any given feature set including clinical features, genomic signatures/profiles, annotated biological pathways, and user-contributed collections of genes. Integrated standard statistical tools provide dynamic quantitative analysis within all available datasets. A new UCSC Cancer Genomics Browser was released July 2012 with immensely enhanced user interface and functionality, new user documentations, and an updated online tutorial.
BioVis 2012 Information