Tractography in Context: Multimodal Visualization of Probabilistic Tractograms in Anatomical Context
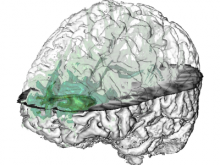
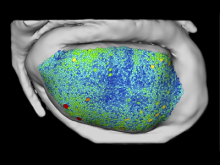
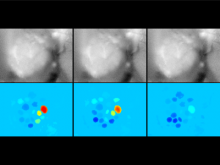
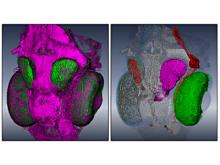
Displaying neurological data in anatomical context is a challenging task in biomedical visualization. We present an application-driven approach solving visibility issues of simultaneously presenting focus and context. \ The tractogram visualization uses nested surface layers with value-based opacity, enhancing shape through depth cues. Anatomical context is given as illustrative, three-dimensional rendering of the cortex, complemented with textured slices. Two user studies were performed to evaluate our methods. \ With our methods, tractogram characteristics become clear, while information and context are perceived simultaneously. They are applicable to a wide range of data as they are easily adaptible to different requirements.
BioVis 2012 Information